vol. 15 no. 1, March, 2010
vol. 15 no. 1, March, 2010 | ||||
In recent years, the biomedical research community, specifically those branches related to human biological repository networks, have been a special target of information sharing efforts. However, very little research has been conducted to investigate the specific information needs of biomedical scientists relevant to the sharing network. These biomedical scientists are a group of scientists whose primarily interest is studying biological functions, phenomena and interaction in the context of medical science. These groups of professionals are unique in terms of the complexity of their multidisciplinary, collaborative research and practices. Traditionally, they have been characterized by their academic disciplines, such as biology, chemistry, physiology and medical science. However, the traditional boundaries of the academic disciplines are increasingly expanding to embrace other fields. In this regard, several biomedical fields have emerged to collaborate with other fields within the sciences for the pursuit of effective and advanced biomedical discoveries.
Increasingly, science, technology, and medical libraries and information centers have worked hard to serve the interdisciplinary group of biomedical professionals in the fast growing genomic era. The human bio-repository network has become critical to genomic research. The network is described as a collaborative resource sharing vehicle for human biological samples (such as human tissues, blood and urine) among multidisciplinary biomedical scientists. The major issue encountered by information centers is the lack of research on the information requirements of biomedical scientists. More importantly, information centres serve their clients within the traditional boundaries of information sources such as journal articles, reference books, monographs, etc. Obviously, biomedical scientists seek not only the scientific discoveries published in academic papers, but also scientific (raw) data such as genomic and proteomic sequences accessible through databases, which are important sources for further references. More demands on non-traditional library collections, such as scientific data, hospital records, clinical images, genomic and proteomic sequences, will challenge libraries and information professionals to understand both the multidisciplinary nature of these scientists and their specific information requirements in various formats.
Obviously, it will be beneficial to biological repositories to learn how the resources are acquired, catalogued, retrieved, and circulated through the library's collaborative approach (e.g., cooperative cataloguing, union collection development, and interlibrary loan). In this way, libraries and information scientists are able to expand their roles by learning the complex nature of biomedical scientists and their information requirements. This study aimed to investigate the characteristics of biomedical scientists and their information requirements, including biological materials as well as data requirements. The understanding of the major characteristics found in this study resulted in a proposed conceptual framework of information requirements for researchers using bio-samples. The framework developed by the current study will enable a system designer to understand such researchers in relation to their professional work roles. More importantly, information scientists will be able to expand their understanding of information organization and the use of physical objects, such as tissues and blood, along with non-traditional information types such as clinical, morphological, and genetic information.
The following sections review three research areas that support the groundwork of this study: biological sample repositories (bio-repositories) and their foremost issues, information requirements of healthcare professionals, and bio-repositories in Korea.
As the demand for biological materials and their associated information increases, there has been a consequent demand for high-quality human bio-samples to support various biomedical researchers worldwide (Mitchell 2000; Gajiwala 2002; Tettamanti et al. 2005; Alvarez et al. 2003; Goebell 2005). The conventional sources for acquiring biological materials provide limited access to those who work in a research environment where an organization's sample collection is only available to in-house researchers. Even if the local collections are available, it is hard to identify the relevant samples for an individual project because of the lack of accompanying sample information. Moreover, the residual samples in the local collection are seldom shared among researchers, many of whom would not have to collect similar samples if a network were available. Potentially, researchers who share similar samples can generate higher validity-based research using a larger sample pool.
To increase accessibility to biological samples, sharing networks have been established. The centralized biological repository does not have to be a physically centralized repository (Eiseman et al. 2003). A virtual network of biological materials accompanied by associated sample information is a more desirable approach (Berman 2003; Gilder 2004; Manley 2001; Lee 2006). The concept of the collaborative sharing network is much like inter-library lending in the library community. It is obvious that all of the required amounts and varieties of samples cannot be supported through a single institution. Geographically diverse locations make the cooperative management of biological samples even harder. In a collaborative research setting, such as multi-centre genetic studies, it is difficult to get approval for the use of biological samples because of varying degrees of local policies on human samples for research. Considering the tremendous amount of research funds and efforts spent for biological samples, it is imperative to construct a centralized repository for a bio-sample resource sharing network (Compton 2005; Friede et al. 2003).
Lack of accessibility to quality human samples is not the only issue for bio-repositories. Most attention has been given to the collection of samples, while no standardized surrogate tool, such as catalogue-like accessibility, has been discussed for the repositories. Repositories range from the commercially available to the federally funded, and more biological samples are accessible through federated online searching tools such as the Specimen Resource Locator, the Tissue Expediter, and National Cancer Institute-Supported Specimen Resources (OBBR) (National Cancer Institute... 2008). However, these tools are limited to a small number of collections on specific disease categories funded by agencies affiliated to the National Institutes of Health. No standardized practice of description has focused on the samples available through repositories. In addition, the information accompanying the collected samples is minimal and can only support limited searching options. It is critical to build an applicable standard for sharing information about quality human samples (Kim and Gilbertson 2007; Kim and Rasmussen 2008).
Additionally, there has been a noticeable dearth of studies on bio-sample users and their data requirements. Various types of biomedical researchers working on complex projects frequently need correlations among the experimental result bio-samples with known parameters for the sample in the experiment. Complex data on each specimen (e.g., sample types, processing, amount, storage, quality assurance and quality control) need to be combined with large numbers of data elements (e.g., pathology data, outcome data and therapy data), and extracted from multiple data sources (e.g., electronic medical records, registries and surgical pathology reports) (Kim and Gilbertson 2007; Kim and Rasmussen 2008). Moreover, the study results for bio-samples (e.g., assay results) are correlated positively to the experimental samples so that the experimental results can be shared among researchers with a similar research interest. As this new area develops and databases or applications are constructed, it is imperative to understand the characteristics of researchers in this field and their data requirements for better resource management.
Drawing upon the contexts of information seeking (behaviour) studies, information requirements by professionals in a specific domain are an important component of an information model. In his seminal review on information behaviour research, Wilson (2000) emphasized that 'the performance of particular tasks, and the processes of planning and decision-making' at work role level is important to information needs. Leckie et al. (1996) found that work roles and tasks are considered prime factors in the information requirements of professionals. Related to the work roles and tasks are professionals' complex job roles involving multiple dimensions of performance (for example, they are expected to perform clinically, scholarly, didactically and managerially). Obviously, the analysis of the information requirements of professionals performing multiple and complex roles is essential to effectively support scientific advancements in biomedical research.
As is evident from previous research findings, few studies have investigated how research-oriented professionals in healthcare seek information to satisfy job-related activities. Most relevant findings can be drawn from studies on basic scientists and engineers. Research done on work-related use of information by basic scientists shows that an information requirement arises from a project, task or problem and the received information affects their productivity and the types of activities they undertake. Studies of diverse professional groups have all concluded that professionals are frustrated in their search for relevant and necessary information (Orr 1970). Frustration becomes more evident if the availability and accessibility of required resources such as bio-samples and the accompanying data are relatively restricted.
In the Orr model, an acceptable timeline and the costs associated with the relevant information are major factors affecting successful information delivery. In Leckie's model, the most important variables are familiarity and prior success with the sources, along with the trustworthiness, packaging, timeliness, cost, quality and accessibility of the sources (Leckie 1996). Therefore, a conceptual framework attempting to capture distinct elements of the information requirement patterns of professionals in the bio-sample domain should be studied for better scientific productivity as well as for effective resource management.
While not widely implemented, there are some data standards for bio-repository information systems which are under extensive review. The Cancer Data Standards Repository, the Cooperative Prostate Cancer Tissue Resources common data element and cancer Text Information Extraction System are currently adopted as data standards by some bio-repository information systems (Patel 2006). It is highly recommended that repositories supported by the National Cancer Institute should be interoperable with core data sets in the Cancer Data Standards Repository (Covitz et al. 2003; McSherry 2008). The College of American Pathologists cancer check lists are also regarded as a pathology findings standard within the cancer Biomedical Informatics Grid semantic framework (College of American Pathologists 2008; Tobias et al. 2006). In addition, the best-practice document of the International Society for Biological and Environmental Repositories specifies the essential data sets to be associated with biological samples (International Society... 2008). These include specimen location, other sample descriptors and additional information for human specimens such as donor information, diagnosis, diagnostic procedures, type of treatment, surgical procedure information, medical history, family history, smoking history, vitals, clinical laboratory values and availability of other biological specimens from the same donor. Likewise, the data elements recommended by various organizations should be compiled into a simple framework for better understanding data and sample requirements. Considering the primitive use of the existing resource sharing networks, the current study was designed to capture pre-analysis of the requirements used for the early stage of system design.
This study is limited to data and sample requirements of Korean biomedical researchers; therefore, the review of the bio-repository network in Korea is briefly described in the following section. Previously, the human bio-repositories in Korea, sponsored by the Korean National Research Resource Centre, have been a major driving force for the systematic collection and storage of biological samples. Currently, more publicly available samples are available through the Centre's network repositories. In the pursuit of a centralized bio-sample network, human repositories such as the Korean Frozen Lung Tissue Bank, Korean Cell Line Bank, Korean Leukemia Cell and Gene Bank, Korean Eye Tissue Bank, Korean Liver Cancer Tissue Bank and a number of branch banks, extend their services to basic and clinical scientists nationwide. For instance, the Korean Frozen Lung Tissue Bank has a virtual tissue network that connects to more than seventeen member branch banks to share human frozen lung tissues and developed comprehensive sample descriptions for advanced searching options. For the broader spectrum of research resource sharing perspectives, the Korean National Research Resource Centre and Singapore Tissue Network, in collaboration with the International Society for Biological and Environmental Repositories, is undertaking discussions to build an Asia-Pacific bio-sample sharing network.
The study was conducted to understand the major characteristics of biospecimen researchers and their bio-sample and data requirements so that biological resources can be effectively and efficiently shared among those who require the resources for scientific discoveries. The following section describes research methods and data analysis performed in the study.
The study addressed the following four research questions:
Several operational definitions are used in this study. 'Information requirements' refers to identified information needs of biomedical scientists that satisfy a work-related goal (modified from Wilson 2000). For studying work-related characteristics, the study used the conventional work roles in bio-repositories, including tissue bankers, medical doctors, and pathology specialists. 'bio-sample-based researchers' are those who require biological samples for their research or work. 'Tissue-bankers' refers to a group of science laboratory workers whose primary responsibilities are to collect, store, annotate and distribute biological samples. 'Pathology specialists' are a more research-oriented group compared to tissue bankers in terms of their research role. The 'medical doctor' group refers to those whose primarily responsibilities are patient care as well as clinical research. The work roles are not exclusive because a pathologist whose primary job is to make a histologic diagnosis for liver cancer may also run a clinical trial research laboratory for discovering effective biomarkers for liver cancer diagnosis. More detailed measures in relation to individual survey questions are discussed below.
Data for this study were obtained from a sample of 137 individuals who answered a request to take part in an online survey between June 19, 2007 and October 19, 2007. The survey was limited to Korean biomedical researchers who were 18 years or older. Invitations to participate in the survey were sent to the Biological Research Information Centre, a major biomedical research message board (http://bric.postech.ac.kr/) and the Korea Human bio-repository Network, a major Korean network. The questionnaire (in Korean) was securely linked to a database in which all information was collected. The institutional review board exemption certificate was obtained before survey distribution. The survey consisted of a self-administered questionnaire and included questions about participants' demographics, research or work-related matters, sample use and requirements and data requirements. The English translation of the questionnaire is included in Appendix A.
The following measures were used to analyse the survey data, including research or work-related matters, sample use and requirement and data requirements. A summary of measures corresponding to survey questions is given in Table 1. The survey asked participants about six different researches or work related activities, including: area of specialties (question 6), funding sources (question 19), interesting organ parts (question 20), interesting diseases (question 22), publication venues (question 23), and attending conferences (question 24). All of these questions target distinct research or work-related activities which can allow us to further assess bio-sample and data requirements. Multiple answers were allowed for those who specialized in more than one area of interest.
| Measure | Survey questions | Measure | Survey questions | Measure | Survey questions |
|---|---|---|---|---|---|
| Research or work-related activities | Area of specialties (question 6) | Sample use and requirements | Human & animal tissue use (questions 7 & 8) | Data equirements | Patient demographics (questions 44-53) |
| Funding sources (question 19) | Experience with bio-repositories (questions 9-11, & 17) | General health conditions (questions 54-60) | |||
| Interesting organ parts (question 20) | Sample types (question 12) |
Laboratory findings (questions 61-67) | |||
| Interesting diseases (question 22) | Processing and storage methods (questions 13 and 14) | Sample-specific elements (questions 68-90) | |||
| Publication venues (question 23) | Requesting amount, frequency and planned budget (questions 15, 16, & 18) | Sample collection (questions 68,70,78-79 & 84) | |||
| Attending conferences (question24) | Request specific criteria (question 21) | Sample processing and storage (questions 69 & 80-82) | |||
| Sample annotation (questions 71-77) | |||||
| Sample distribution (questions 86-90) | |||||
Sample use and requirements were assessed using thirteen questions, which asked respondents about their experiences of bio-sample usage and current or expected sample requirements, including human and animal tissue use (question 7 & 8); experience with bio-repositories (questions 9-11, & Q17); sample types (question 12); processing and storage methods (questions 13 and 14); requesting amount, frequency, and planned budget (questions 15, 16 & 18); and request specific criteria (question 21). The response options were partially modified from caTISSUE Core search fields (cancer Biomedical Informatics Grid tissue bank repository tool) and National Biospecimen Network Blueprint survey questions on bio-sample and data requirements (Kim and Rasmussen 2008; Friede et al. 2003; McSherry and Paul 2008). The caTISSUE Core was used in this study because it is the open source recommended by the National Cancer Institute for wide usage by comprehensive cancer centers in the U.S.
A list of required data elements to assess the participants' needs for specific kinds of information was collected from three sources, including the (US) Health Insurance Portability and Accountability Act (HIPAA) identifiers, Korean Frozen Lung Tissue Bank and caTISSUE Core, which are considered the representatives of protected and usable data elements regulated or recommended by government-affiliated agencies. A measure for data requirements was further divided into HIPAA protected identifiers as opposed to non-HIPAA elements. The HIPAA elements were included in the current study because it is original source for the Korean Bioethics regulation. The HIPAA-regulated data elements exist to protect patients' private and confidential information. Since human biological samples are subject to the HIPAA rule, it is meaningful to assess whether or not the requested data elements are HIPAA indicators or not. The non-HIPAA data elements were then further grouped into demographics (questions 44-53), general health conditions (questions 54-60), lab findings (questions 61-67), and sample-specific elements (questions 68-90). In addition, the study did not intend to exhaustively cover the entire spectrum of data elements required for sample annotation, rather to survey core annotation elements based on the existing sources such as the caTISSUE Core and Korean Frozen Lung Tissue Ban system. The sample-specific questions were also analysed into sub-activities of repositories, such as collection (questions 68, 70, 78-79 &84), processing and storage (questions 69 & 80-82), annotation (questions 71-77), and distribution (questions 86-90).
The study used three data analyses including descriptive analysis, cross-tabulation, and the TwoStep Cluster analysis for the research questions. SPSS (version 15) statistical software was used for the statistical analyses. For research question 1, the study used descriptive statistics to characterize the participants based on the survey data collected. Cross-tabulation was also performed to compare different groups of participants including the medical doctor group (N=11, 8%), pathology specialists (N=33, 24.1%), and tissue bankers (N=37, 27%) for their bio-samples and data requirements. The TwoStep Cluster analysis was used to assess whether there were any natural groups of the participants whose sample and data requirements were distinct from one another. The TwoStep Cluster method was chosen because the algorithm was built to handle mixed data types (continuous, binary, categorical, etc.) (SPSS Inc., 2008). Both continuous and categorical variables (or attributes) were used to calculate similarity (or dissimilarity) to identify any natural groups. The first step was to pre-cluster the cases (or records) into many small sub-clusters. The desired number of clusters resulting from the pre-cluster step were determined if specific numbers were not defined.
The study sought to describe the survey respondents by assessing general demographic characteristics, work or research-related characteristics, bio-sample use and requirement-related characteristics, and data requirement characteristics. 137 respondents who replied to the survey between July 19, 2007 and October 19, 2007 were included in the study. Of these, 114 (83.2%) were human sample users and 89 (65%) were animal sample users. Sixty-six respondents used both human and animal samples. Ninety-seven participants (71%) were and 28 (21%) were female. Ten people (8%) did not answer. Eleven participants (8%) had medical doctorates were and 33 (c.24%) identified themselves as pathology laboratory medicine specialists. Thirty-seven participants (27%) answered that they had experience working at a bio-repository.
The sample was not equally distributed among medical doctorates, pathology specialists and tissue bankers, which may have an impact on the study results. The most prevalent age group found in this survey was between 26 and 35, representing relatively junior researchers. 'Years of work experience' also showed that the survey respondents were new entrants to the field of biomedical sciences. Abridged descriptive survey results are shown in Table 2.
| Measure | Result | Freq. (%) | Measure | Result | Freq. (%) |
|---|---|---|---|---|---|
| * Specialty (question 6) | Molecular biology | 14 (10.2) | Human sample (question 7) | Yes | 114 (83.2) |
| Genetics | 26 (19.0) | Animal sample use (question 8) | Yes | 89 (65) | |
| Pathology | 33 (24.1) | Repository use (question9) | Yes | 44 (32.1) | |
| Biochemistry | 12 (8.8) | Repository work (question 10) | Yes | 37 (27) | |
| Public health | 54 (39.4) | * Types of repository work (question 11) | Sample collection | 28 (20.4) | |
| Biostatistics | 5 (3.6) | Sample processing | 20 (14.6) | ||
| Health administration | 1 (0.7) | Sample storage | 27 (19.7) | ||
| * Diseases (question 22) | Liver and digestive system | 27 (19.7) | Sample distribution | 13 (9.5) | |
| Musculoskeletal & connective tissue | 17 (12.4) | Information management | 17 (12.4) | ||
| Neoplasms (lung, breast, ovary) | 26 (19) | Ethical, legal, administration | 12 (8.8) | ||
| Cardiac-related | 19 (13.9) | * Sample types (question 12) | Molecular | 89 (65) | |
| Infectious and parasitic | 32 (23.4) | Cell | 62 (45.2) | ||
| Diabetes and endocrine-related | 19 (13.9) | Fluid | 80 (58.4) | ||
| Others | 52 (38) | Tissue | 72 (52.6) | ||
| Data elements required | HIPAA elements (questions 25-43) | 2.04 (1.28) | * Sample requesting criteria (question 21) | By anatomic sites | 61 (44.5) |
| Demographics (question 44-53) | 2.99 (1.37) | Either by normal or diseased | 76 (55.5) | ||
| General health conditions (questions 54-60) | 3.63 (1.35) | By diseases | 75 (54.7) | ||
| Laboratory findings (questions 61-67) | 3.58 (1.31) | By primary or metastatistic | 44 (32.1) | ||
| Sample-specific elements (questions 68-90) | 3.75 (1.24) | By matched normal from the same patient | 35 (25.5) | ||
| Sample collection (questions 68,70,78-79 & 84) | 3.83 (1.26) | By specific treatments on patients | 30 (21.9) | ||
| Processing & storage (questions 69 & 80-82) | 3.60 (1.25) | By tissue sources | 19 (13.9) | ||
| Sample annotation (questions 71-77) | 3.86 (1.27) | By demographic condition | 36 (26.3) | ||
| Sample distribution (questions 86-90) | 3.59 (1.18) | By amount of tissue | 38 (27.7) | ||
| By total number of specimens needed | 31 (22.6) | ||||
| By specimen preparation and preservation method | 47 (34.3) |
In terms of work and research related characteristics, the most reported anatomic sites of interests included lymphatic and immune related (N=45, 32.85%), endocrine (N=29, 21.17%), and stem cells and reproductive organs (N=29, 21.7%). In addition, the respondents replied that the most interesting research focus of the diseases were infectious and parasitic-related (N=32, 23.4%), followed by neoplasms (N=26, 19%). The survey respondents listed eighty two unique journal titles as potential publication places and 115 unique professional and academic conferences implying the respondents' involvement in specialized research areas.
The study also asked about sample specific requirements. With slightly over a decade of bio-repository experience in Korea, over 32 percent of the participants (N=44) answered that they used bio-repositories to acquire bio-samples. The majority of the participants also answered that more than one sample type was required for their individual project. Highly demanded types of bio-samples included RNA, cryopreserved cells, serum/whole blood, and frozen samples. In addition, PCR and RT-PCR were identified as the most required sample processing methods. The majority of the respondents also answered that they would like to have the sample stored in fresh-frozen, frozen, and paraffin blocks. Questions regarding sample requesting criteria identified that the most prevalent search criteria included search by anatomic sites, either by normal or diseased, or by disease names. These findings are consistent with previous studies (Kim and Gilbertson 2007; Kim and Rasmussen 2008).
Questions asked about data requirements were further analysed according to Health Insurance Portability and Accountability Act-restricted data elements, demographic data elements, general health condition data elements, laboratory finding elements, and sample specific data elements. The study found that the personally identifiable information that is under restricted distribution by the HIPAA and the Korean Bioethics and Biosafety Law were not frequently requested data elements by the respondents. More importantly, sample specific data such as sample diagnosis, processing, storage medium and condition, were highly demanded by the respondents (Avg=3.75, Std=1.24). These data indicate that researchers require more sample-specific data when they request bio-samples rather than personally identifiable data elements. Laboratory and physiology findings were also considered important data followed by general health condition and patient health history. As shown in the 2003 National Biospecimen Network survey, more than a single source was identified by the respondents as required data to be accompanied with bio-samples (Friede et al. 2003). This implies that frequently required data are complex in nature and cannot be extracted from a single medical information system.
The study intended to characterize participants by their work roles representing medical doctors, pathology specialists, and tissue bankers. The categorization of work roles was not mutually exclusive; therefore, one person could belong to more than one work-related category. These categories were then used to further characterize the study participants in terms of bio-sample and data requirements.
| Samples requirements | Medical doctor (N=11) | Non-medical doctor (N=126) | Pathology (N=33) | Non-pathology (N=104) | Tissue banker (N=37) | Non-tissue banker (N=100) |
|---|---|---|---|---|---|---|
| Freq. (%) | Freq. (%) | Freq. (%) | Freq. (%) | Freq. (%) | Freq. (%) | |
| Human sample use | 11 (100) | 103 (75.2) | 33 (100) | 81 (77.9) | 34 (91.9) | 80 (70.2) |
| Animal sample use | 8 (81.8) | 81 (81.7) | 16 (48.5) | 73 (70.2) | 24 (64.9) | 65 (73) |
| Repository used | 5 (45.5) | 88 (69.8) | 11 (33.3) | 33 (31.7) | 16 (43.2) | 28 (63.6) |
| # Requesting sources: -Pathology department -Research laboratory | 6 (54.5) 1 (9.1) | 45 (35.7) 47 (37.3) | 21 (63.6) 5 (15.2) | 30 (28.8) 43 (41.3) | 12 (32.4) 7 (18.9) | 39 (39) 41 (41) |
| # Sample Type: -RNA -Cryopreserved cells -Serum/whole blood -Frozen | 9 (81.8) 5 (45.5) 5 (45.5) 8 (72.7) | 80 (63.5) 57 (63.5) 75 (59.5) 64 (50.8) | 21 (63.6) 14 (42.4) 15 (45.5) 20 (60.6) | 68 (65.4) 50 (48.1) 65 (62.5) 52 (50) | 25 (67.6) 24 (64.9) 21 (56.8) 22 (59.5) | 64 (64) 41 (41) 59 (59) 50 (50) |
| # Sample processing: -PCR -RT-PCR |
8 (72.7) 7 (63.6) |
92 (73) 91 (72.2) |
25 (75.8) 22 (66.7) |
75 (72.1) 76 (73.1) |
27 (73) 30 (81.1) |
73 (73) 68 (68) |
| # Sample storage: -Fresh-frozen -Frozen -Paraffin |
8 (72.7) 5 (45.5) 8 (72.7) |
59 (46.8) 66 (52.4) 42 (33.3) |
17 (51.5) 16 (48.5) 17 (51.5) |
50 (48.1) 55 (52.9) 33 (31.7) |
20 (54.1) 19 (51.4) 16 (43.2) | 47 (47) 52 (52) 34 (34) |
| Amount required | 22g/y | 50g/y | 15g/y | 58g/y | 15g/y | 59g/y |
| Frequency required | 2.18/y | 29/y | 30.68/y | 26.16/ | 44.1/y | 21.02/y |
| Price willing to pay | $430 | $500 | $342 | $542 | $534 | $478 |
| # Anatomic sites: -Immune systems related -Respiratory |
0 4 (36.4) |
45 (35.7) 9 (7.1) |
5 (15.2) 6 (18.2) |
40 (38.5) 7 (6.7) |
6 (16.2) 3 (8.1) |
39 (39) 10 (10) |
| # Diseases of interest: -Liver/digestive -Immunology-related -Cancer-related |
1 (9.1) 1 (9.1) 5 (45.5) |
26 (20.6) 31 (24.6) 21 (16.7) |
3 (9.1) 4 (12.1) 8 (24.2) |
24 (23.1) 28 (26.9) 18 (17.3) |
7 (18.9) 8 (21.6) 9 (24.3) |
20 (20) 24 (24) 17 (17) |
| Requesting criteria: -By anatomic sites -Normal or diseased -By disease -Primary or metastatic -Matched normal -By treatment -Tissue sources -By demographic -By available amounts -By available numbers -By sample preparation |
7 (63.6) 5 (45.5) 7 (63.6) 6 (54.5) 4 (36.4) 4 (36.4) 2 (18.2) 4 (36.4) 4 (36.4) 5 (45.5) 6 (54.5) |
54 (42.9) 71 (56.3) 68 (54) 38 (30.2) 31 (24.6) 26 (20.6) 17 (13.5) 32 (25.4) 34 (27) 26 (20.6) 41 (32.5) |
14 (42.4) 16 (48.5) 18 (54.5) 14 (42.4) 7 (21.2) 9 (27.3) 2 (6.1) 9 (27.3) 6 (18.2) 6 (18.2) 11 (33.3) |
47 (45.2) 60 (57.7) 57 (54.8) 30 (28.8) 28 (26.9) 21 (20.3) 17 (16.3) 27 (26) 32 (30.8) 25 (24) 36 (34.6) |
16 (43.2) 20 (54.1) 22 (59.5) 15 (40.5) 10 (27) 9 (24.3) 6 (16.2) 13 (35.1) 10 (27) 11 (29.7) 16 (43.2) |
45 (45) 56 (56) 53 (53) 29 (29) 25 (25) 21 (21) 13 (13) 23 (23) 28 (28) 20 (20) 31 (31) |
| Journals -Clinical -Basic science -Unknown |
6 (54.5) 3 (27.3) 4 (36.4) |
37 (29.4) 50 (39.7) 58 (46) |
11 (33.3) 7 (21.2) 17 (51.5) |
32 (30.8) 46 (44.2) 45 (43.3) |
16 (43.2) 15 (40.5) 14 (37.8) |
27 (27) 38 (38) 48 (48) |
| Professional associations -Clinical -Basic science -Unknown |
7 (63.6) 2 (18.2) 2 (18.2) |
37 (29.4) 72 (57.1) 29 (23) |
15 (45.5) 7 (21.2) 8 (24.2) |
29 (27.9) 67 (64.4) 23 (22.1) |
18 (48.6) 13 (35.1) 8 (21.6) |
26 (26) 61 (61) 23 (23) |
As shown in Table 3, all medical doctor and pathology respondents (100%) replied that they used human tissues, while slightly over 70 percent of other groups used human samples. Compared to other groups, fewer pathology respondents used animal sample users (N=16, 48.5%). More pathology and medical doctor respondents identified that they requested bio-samples through pathology rather than other bio-sample sources. This implies that easier accessibility to traditional bio-repository such as pathology is given to clinical groups compared to non-clinical groups. For sample processing and sample type, a majority of medical doctors answered that RNA (N=9, 81.8%) and frozen samples (N=8, 72.7%) were the most demanded samples, while other groups demanded other types of samples equally. It is also interesting to note that non-medical doctors, non-pathology, and non-tissue bankers reported that they required a larger amount of human samples and were less willing to pay for use of human samples compared to other groups. Cancer-related research was the most frequent disease of interest identified by the medical doctor group (N=5, 45.5%). More clinically oriented journals were identified as potential publication places by medical doctors compared to other groups (N=6, 54.5%). This result was consistent in clinically oriented conferences and associations that medical doctors and pathology groups normally attend.
Data requirements by different groups are shown in Table 3. The highest data requirement score reported for the Health Insurance Portability and Accountability Act-related data was demanded by non-tissue bankers (Mean=2.07, Standard deviation=1.31) followed by non-pathology (Mean=2.06, Standard deviation=1.29) and non-medical doctors (Mean=1.99, Standard deviation=1.26). More general demographics about patients were reported as important data for medical doctors (Mean=3.06, Standard deviation=1.43) compared to other groups. Important scores of data requirements by different groups were distinct in health history elements. Medical doctors (Mean=3.97, Standard deviation=1.28), pathology (Mean=3.83, Standard deviation=1.27), and tissue bankers (Mean=3.90, Standard deviation=1.19) were more likely to require health history than other groups. For laboratory findings, there were slightly higher scores reported by tissue bankers (Mean=3.66, Standard deviation=1.17), however, the difference was not significant. For sample specific requirements, medical doctors (Mean=3.89, Standard deviation=1.28), pathology (Mean=3.75, Standard deviation=1.22), and tissue bankers (Mean=3.80, Standard deviation=1.18) scored slightly more than other groups.
| Respondents' data requirements | Medical doctor Mean (SDev.) |
Non-medical doctor Mean (SDev.) |
Pathology Mean (SDev.) |
Non-pathology Mean (SDev.) |
Tissue banker Mean (SDev.) |
Non-tissue banker Mean (SDev.) |
|---|---|---|---|---|---|---|
| HIPAA | 1.74 (1.34) | 1.99 (1.26 ) | 1.67 (1.12) | 2.06 (1.29) | 1.70 (1.07) | 2.07 (1.31) |
| Non-HIPAA | 3.68 (1.28) | 3.53 (1.29) | 3.57 (1.26) | 3.53 (1.29) | 3.61 (1.22) | 3.51 (1.31) |
| Demographics | 3.06 (1.43) | 2.98 (1.36) | 2.91 (1.40) | 3.02 (1.35) | 2.93 (1.37) | 3.01 (1.36) |
| Non demographic | 3.18 (1.28) | 3.14 (1.26) | 3.08 (1.19) | 3.15 (1.28) | 3.11 (1.15) | 3.14 (1.30) |
| Health history | 3.97 (1.28) | 3.60 (1.34) | 3.83 (1.27) | 3.57 (1.36) | 3.90 (1.19) | 3.53 (1.38) |
| Non-health history | 3.05 (1.30) | 3.05 (1.27) | 2.96 (1.22) | 3.08 (1.28) | 2.99 (1.18) | 3.07 (1.30) |
| Laboratory findings | 3.52 (1.10) | 3.58 (1.31) | 3.66 (1.17) | 3.55 (1.32) | 3.59 (1.20) | 3.58 (1.33) |
| Non-laboratory finding | 3.10 (1.32) | 3.05 (1.27) | 2.92 (1.23) | 3.03 (1.29) | 3.03 (1.18) | 3.07 (1.31) |
| Sample specific | 3.89 (1.28) | 3.71 (1.23) | 3.75 (1.22) | 3.72 (1.24) | 3.80 (1.18) | 3.70 (1.26) |
| Not-sample specific | 2.72 (1.31) | 2.76 (1.30) | 1.94 (2.00) | 2.05 (1.65) | 2.67 (1.18) | 2.79 (1.34) |
The study assessed whether there are any natural groups of the participants whose bio-sample and data requirements are similar within groups. A complete summary of TwoStep Cluster results can be found in Table 5. TwoStep Cluster analysis resulted in two distinct groups including Cluster 1 (N=116) and Cluster 2 (N=21). Cluster 1 includes the majority of specialty groups (medical doctors, pathology, and tissue bankers), while Cluster 2 include more non-specialty groups (non-medical doctors, non-pathology, and non-tissue bankers). These data imply that the respondent category used in this study is helpful in grouping bio-sample users which can be implemented in modelling distinct work roles for bio-sample-related users.
| Respondents' variables | Cluster 1 (N=116, 84.67%) | Cluster 2 (N=21, 15.33%) | |
|---|---|---|---|
| Respondent category | Medical doctors (N=11) | 10 (90.91) | 1 (9.09) |
| Pathology (N=33) | 30 (90.91) | 3 (9.09) | |
| Tissue bankers (N=37) | 33 (89.19) | 4 (10.81) | |
| Sample requirements | Human sample used | 101 (88.59) | 13 (61.9) |
| Animal sample used | 76 (65.51) | 13 (61.9) | |
| Repository used | 38 (32.75) | 6 (28.57) | |
| # Requesting sources: -Pathology -Other Labs |
45 (38.79) 40 (34.48) |
6 (28.57) 8 (38.10) |
|
| Sample Type -RNA -Cryopreserved cells -Serum or whole blood -Frozen |
77 (66.38) 53 (45.69) 70 (60.34) 62 (53.45) |
12 (57.14) 9 (42.86) 10 (47.62) 10 (47.62) |
|
| Sample processing -PCR -RT-PCR |
85 (73.28) 85 (73.28) |
15 (71.43) 13 (61.9) |
|
| Sample storage -Fresh-frozen -Frozen -Paraffin |
60 (52.59) 61 (53.45) 44 (37.93) |
7 (33.33) 10 (47.62) 6 (28.57) |
|
| Amount required | 54g/year | 11g/year | |
| Frequency required | 25.89/year | 34.76/year | |
| Price willing to pay | $502 | $448 | |
| Anatomic sites: -Immune systems related -Respiratory |
38 (32.75) 13 (11.21) |
7 (33.33) 0 |
|
| Diseases of interest: -Liver or digestive system -Immunology-related -Cancer-related |
53 (45.69) 67 (57.76) 65 (56.03) |
8 (38.10) 9 (42.86) 10 (47.62) |
|
| Requesting criteria: -By anatomic sites -Normal or diseased -By disease -Primary or metastatic -Matched normal -By treatment -Tissue sources -By demographic -By available amounts -By available numbers -By sample preparation |
21 (18.10) 13 (11.21) 8 (6.90) 13 (11.21) 26 (22.31) 22 (18.97) 8 (6.90) 17 (14.66) 10 (11.6) 7 (5.56) 10 (11.6) |
6 (28.57) 4 (19.05) 1 (4.76) 6 (28.57) 6 (28.57) 4 (19.05) 2 (9.52) 2 (9.52) 2 (9.52) 1 (4.76) 3 (14.29) |
|
| Journals -Clinical -Basic science -Unknown |
37 (31.90) 42 (36.20) 54 (46.55) |
6 (28.57) 11 (52.38) 8 (38.10) |
|
| Associations -Clinical -Basic Science -Unknown |
37 (31.90) 61 (52.59) 27 (23.28) |
7 (33.33) 13 (61.9) 4 (19.05) |
|
| Data requirements | HIPAA | 1.92 (1.18) | 1.97 (1.26) |
| Non-HIPAA | 3.75 (1.09) | 3.48 (1.31) | |
| Demographics | 3.17 (1.26) | 2.99 (1.36) | |
| Non-demographic | 3.44 (1.07) | 3.23 (1.28) | |
| Health history | 3.92 (1.11) | 3.63 (1.34) | |
| Non-health history | 3.25 (1.10) | 3.07 (1.29) | |
| Laboratory findings | 3.88 (1.06) | 3.58 (1.29) | |
| Non-laboratory finding | 3.26 (1.12) | 3.08 (1.30) | |
| Sample specific | 4.05 (0.93) | 3.73 (1.24) | |
| Not-sample specific | 3.22 (1.15) | 3.04 (1.31) | |
For use of bio-samples, a larger portion of bio-sample users were clustered in Cluster 1 (N=101, 88.59%) than Cluster 2 (N=13, 61.9%). A higher percentage of people in Cluster 1 (N=38, 32.75%) used bio-repositories compared to Cluster 2 (N=6, 28.57%). About 39 percent of Cluster 1 (N=45) requested bio-samples through pathology, while only less than 30 percent of Cluster 2 (N=6) requested bio-samples through pathology. Heavy bio-sample users requiring greater amounts and more frequently required bio-samples were clustered in Cluster 1 than Cluster 2. Cluster 1 indicated more requesting criteria when searching bio-samples than Cluster 2. These indicate less demands of the associated bio-sample data by Cluster 2 compared to Cluster 1. Distinct characteristics between clinical and non-clinical groups were also found in these two clusters. Cluster 1 is more likely clinically oriented compared to Cluster 2 whose publication places and professional associations (and conferences) were more likely basic science-oriented.
Some distinct characteristics between the two clusters were reported based on the importance scores of the various data elements given. For almost every data requirement, Cluster 2 scored higher than Cluster 1. Only the non-HIPAA-related data group was slightly more demanded by Cluster 2 (Avg=1.97, Std=1.26) compared to Cluster 1 (Avg=1.92, Std=1.18). Sample-specific data requirements (Avg=4.05, Std=0.93) were the most highly scored data requirement by Cluster 1 followed by health history (Avg=3.92, Std=1.11). The highest scored data requirement by Cluster 1 was sample specific (Avg=3.73, Std=1.24) data. Least scored elements by both Cluster 1 (Avg=1.92, Std=1.18) and Cluster 2 (Avg=1.97, Std=1.26) were HIPAA restricted data elements. Overall, Cluster 2 included active and heavy bio-sample users whose members were more clinically oriented compared to Cluster 1. This result supports previous research findings about two distinct bio-sample user groups including clinically oriented (Cluster 1) and basic science-oriented groups (Cluster 2).
The proposed conceptual framework of information requirements of bio-sample using researchers based on the current study findings and the reviews of the previous studies was developed. A basic framework to develop the current study model was adopted from Leckie's model (1996). A complete model is depicted in Figure 1. The basic components of the proposed model are the professional work roles and their related tasks performed by tissue users in their daily practice. In addition, interacting components of information seeking included characteristics of data and bio-sample needs and factors affecting information seeking (sources and awareness) which resulted in the outcome component. Individual boxes in Figure 1 represent each core component of the model. The model proposed here is not intended to be comprehensive but is a conceptual model that can be further tested and refined in a larger empirical study. The following section will discuss key components contained in the proposed framework.
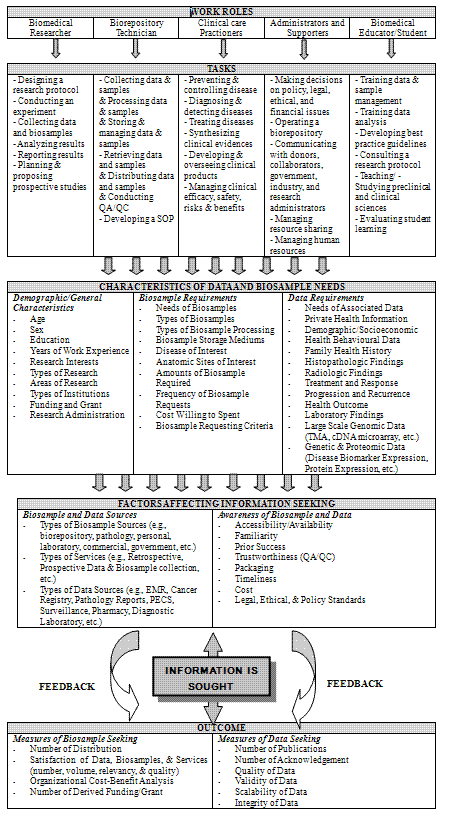
As found in various empirical studies, professionals work in complicated environments and play complex roles in regard to information seeking. The first box of Figure 1 lists five professional roles found in the current study and previous research: biomedical researchers, bio-repository technicians, clinical care practioners, administrators and supporters, and biomedical educators and students. The individual roles of tissue users are highly related to their professional tasks described in the tasks section in Figure 1. For instance, the main tasks of biomedical researchers are to design and conduct studies, collect and analyse data, report findings and plan for further studies. These roles are not different from any other researchers. bio-repository technicians, who were identified as tissue bankers in the current study, play important roles in collecting, processing, storing, retrieving, and distributing bio-samples and their related data which are also linked to quality assessment for the collected samples and data. Clinical practitioners who were identified as medical doctors and pathologists focus on clinical care; therefore, their professional roles are more relevant to prevention, diagnosis, treatment, prognosis and management of clinical care. Administrators and supporters are professionals whose roles focus on making managerial decisions, operating the bio-repositories, building external and internal networks, and improving efficacy, safety, minimizing risks and maximising benefits of the bio-repository. The student group is assigned in the last user role along with biomedical educators. The majority of the respondents to the current study were junior researchers who were still in training. Their bio-samples and the associated data requirements should not be neglected.
There are also other general characteristics of information needs that enter into the component of the proposed model. The study identified three distinct characteristics of information requirements: demographic and general characteristics, bio-sample requirements and data requirements. Demographic and general characteristics are components that describe individual users. The current study's survey questions used individual characteristics along with the questions asked about bio-sample specific requirements and data specific requirements. The data findings and bio-sample requirements listed in the third box of Figure 1 are the core of the information seeking model because these findings directly relate the information required to those who request the information.
Factors affecting information seeking are listed in the fourth box in Figure 1. Numerous information seeking studies of professionals have sought to determine what factors affect information seeking behaviour. In the current study, sources of bio-samples and data were identified as a major factor in users seeking information from a number of bio-sample and data sources including bio-repositories, pathology laboratories, personal laboratories, electronic medical records, surgical pathology reports and cancer registries. The study also included awareness of bio-sample and data, which may affect information seeking. The identified factors should be further explored to assess whether a certain factor might have influence on other factors (compounding influence on multiple factors).
In the last information seeking process, outcome, is the 'end point of the work-related requirements of specific roles and tasks' (Wilson 2000). The study identified measurements of bio-sample seeking and data seeking. These measures should be further developed so that quality assurance and successful data and bio-sample distribution may be scientifically measured. The number of sample distributions, satisfaction with data and bio-sample related services, organizational cost-benefit analysis, and the amount of derived funding and grants as well as the number of publications can be used as measures of the outcome. More importantly, feedback should be carefully conducted so that information seeking does not flow in one direction, but is an interactive process based on the identified needs of information seekers.
The study was designed to describe the general characteristics of biomedical scientists who require human biological samples and associated data. The findings uncovered specific requirements of the physical bio-samples and the related data from multiple information systems. The results reported here are not directly indicative of what science, technology, and medical libraries should acquire, annotate, circulate and serve for their current clientele. Rather, this study should lead to the discussion of new types of information sources (e.g., human tissues and blood), the complex nature of the information seeking behaviour of biomedical scientists (and, in particular, those using bio-samples) and the expanded boundary of information service centres such as bio-repository libraries. The following discussion of the study findings highlights the data presented in the previous section.
First, the finding of distinct characteristics for different professional roles in seeking biological samples and associated data will be beneficial to the development of a framework for information seeking behaviour. In a previous study, Kim and Gilbertson found two distinct user groups described by their sample and data requirements. The current study also confirmed that there were distinct characteristics among two clusters of users. One group (C1) was more likely to be clinically-oriented than the other group (C2). For instance, the clinical group has easier access to human samples compared to the non-clinical group, so the sample requirements and requesting criteria reported are diverse and detailed. Not only the sample requirements but also the data requirements confirmed that the basic science group (C2) is less discriminating regarding their choice of data requesting criteria (e.g., by anatomic sites, normal and matched abnormal, and primary and metastatic samples) (Kim and Gilbertson 2007). This implies that there will be potential benefit for basic scientists to develop more elaborate sample-specific study variables if the data are accessible through a well-managed bio-repository database. Likewise, more elaborate efforts should be made to identify clinically relevant genomic variables to be recorded with human tissue samples.
Secondly, the current study found that tissue users were mostly junior investigators from multiple academic disciplines conducting various types of basic, developmental, translational, and clinical research and they were largely from academic medical centres whose interests in anatomic sites and diseases varied. This finding supports the growing trends towards biomedical research requiring human samples in both clinical and basic sciences among relatively new investigators, whose bio-sample requirements are to be supported through a systematic management of bio-repositories. The wide variety of sample and data requirements found in this study is a strong indicator of the necessity of a nationwide network to serve biomedical researchers with less accessibility to human biological samples. For instance, if a junior investigator wants to demonstrate the potential of biomarkers in the development of a cancer drug such as Trastuzumb, a recombinant monoclonal antibody in Asian populations, could benefit by accessing biomarker-based patient selection at an early stage in the clinical trial process, which could optimize the development of successful cancer therapy (Friede et al. 2003). Through a biological resource sharing network, researchers from non-clinical disciplines can also access valuable resources at lower cost.
Thirdly, the current study set out to discover complex and multidisciplinary biomedical researchers whose information seeking not only focuses on textual data but also on physical items that have not been given much attention by information scientists. The study learned that managing physical bio-samples is very different from managing conventional, textual information sources. For instance, various sample-specific requirements should be tracked, including sample location, condition, availability, history and significant events (such as sample thaws, loss, destruction and processing of any kind), as well as specimen distribution through a unique identifier which can be also linked to other established databases. Frequent updates are required for most of the sample-specific information, especially when the samples are dispensed. This means that physical objects, such as human tissues, are not expected to be returned for continued use. For this reason, some tissue banks recommend researchers deposit the study results (data generated using the distributed samples) rather than the physical tissues. So, the designer of the bio-repository system should anticipate the resulting scientific data to be stored, retrieved and redistributed for further use. In other words, once a library holding (e.g., a tissue sample) is circulated to a researcher, the library should plan for acquiring the end result as a reciprocal benefit. This will ensure that other researchers do not have to repeat a study with the same samples, if the study results are accessible to them. Designing information systems that manage biological samples and the multi-faceted biomedical data is not easy task for information scientists who are used to conventional bibliographic management systems. Although the findings are limited to the survey respondents, the results can be used as representative use cases with which to construct bio-sample research databases.
Most importantly, the study findings indicate that a standardized information set for sample collection, processing methods and storage conditions should be available to the user to aid in selecting specimens for testing. Along with the relevant biomedical data, which are to be retrieved from multiple information sources, all of the sample specific requirements ought to be accessible through a standardized data format. For instance, polymerase chain reaction amplification techniques require only a few nanograms, which can be served by a milligram of tissue left over from standardized diagnostic procedures. For further advanced techniques, such as specimens for whole-proteome analysis or microarrays for whole-genome analysis, it is advisable to develop a strategic plan to save tissue for future use when more advanced procedures become available.
Standardization is significant not only for physical samples, but also for relevant data. As the study findings suggest, the respondents extensively require various bio-sample-related data. As the International Society for Biological and Environmental Repositories Best Practice (2008) recommends, information accompanying biological samples should cover a varying degree of data which can be either linked to an external database or directly linked to the repository information system. For instance, information regarding specimen location, status, condition, collection, processing, storage and distribution was found to be relevant data to the study respondents. Considering the nature of human biological samples, additional information regarding patient-specific data such as age at the time of collection, sex, occupation, race or ethnicity, diagnosis, diagnostic procedure, types of treatment, surgical procedure, medical and family history, and other health behavioural information such as smoking and nutrition, physiological and clinical laboratory data, and availability of other biological samples would be beneficial to the biomedical researcher who might not consider combining clinical data with genomic data (single nucleotide polymorphisms, mutations, microarrays) and proteomic data (specific protein biomarkers, two-dimensional gel data, mass spectral analyses). Obviously, the information stored will vary depending on the nature, purpose, and intended uses for the biological collection; however, careful consideration should be given to what the repository information system should or can contain depending on who the potential users are. As discussed in numerous behaviour studies, information that is easily accessible is used more by scientists (Kim 2009). Therefore, the challenge for information scientists is to construct an effective and efficient information vehicle through which biomedical scientists can easily identify samples along with relevant data.
Lastly, the major contribution of the current study is the development of an information requirement model based on researchers use of biological samples and their related data requirements. The characteristics and variables developed in the current study can be used for future empirical studies assessing potential users, types of information required depending on work-related roles, factors affecting information seeking, and evaluation of information seeking effectiveness. The current study found that information defined in the tissue-related model is different from Leckie's generic model. Information required by these groups is not only a physical entity such as tissues, serum, plasma and molecular material, but also the entity's associated data extracted from various clinical and research databases such as electronic medical records, surgical pathology reports, cancer registry and genome analysis information systems. Likewise, the proposed model stresses the requirements of bio-samples and their associated data that make tissue users distinct from other professionals. Therefore, the growing trend toward the systematic management of bio-samples and relevant data, are to be carefully studied for effective resource sharing.
The study formed a framework to capture the information seeking behaviour of researchers using bio-samples. This framework introduces a greater understanding of the professional roles and tasks, which relates both directly and indirectly to the use of bio-samples and data. The findings show that the information requirements of bio-sample-related researchers are heavily influenced by the professional role-task relationship. Therefore, general factors such as researchers' demographics, professional career stages, or recurring needs should be carefully explored in relation to the role-task relationship for further study (Fidel and Green 2004; Kari 2009; Bellazzi and Zupan 2008). Moreover, a noticeable dearth of information has been reported in the area of predictive data mining in clinical medicine where limited use of genomic and proteomic findings were included to develop a clinical outcome prediction model (Gilbertson et al. 2004).
One of the limitations of the current study is that the concept of information seeking behaviour (Leckie's) was used to model a framework. Leckie's model was built to understand seeking and behaviour rather than information requirements in an early stage of system development. Although it is the closest and the most relevant information model of professionals, it may not be perfectly suited to the current study. So, the findings will be more beneficial to designing an information system than to understand the overall information-seeking behaviour of scientists (Meho and Tibbo 2003). Additionally, the survey was limited to Korean researchers and should be expanded to researchers other than Koreans so that the study results are more representative of other populations.
In conclusion, bio-sample resource sharing and distribution will not be successful unless both the physical integrity of the biomolecules and the value-added information associated with the sample are made accessible through a formal information requirement analysis (Schilsky et al. 2002; Hu et al. 2004; Hanson 1998). In order to expand the roles of information professionals in science, technology, and medicine, the following must be clearly understood in the emerging genetic era: Who needs what information why and when, and in order to accomplish what tasks?
This project was supported in part by a faculty research grant by college of communication and information studies, University of Kentucky. In addition, this publication was made possibly by Grant Number P20RR-16481 from the National Centre for Research Resources, a component of the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of National Centre for Research Resources or National Institutes of Health.
Dr. Sujin Kim, is an assistant professor in the School of Library and Information Science and is jointly appointed to the Department of Pathology and Laboratory Medicine, University of Kentucky, Lexington, Kentucky, USA. She was trained in the pathology and oncology informatics group at the University of Pittsburgh, USA, working with microscopic images and human bio-sample repositories. Her specialty is biomedical informatics and health science librarianship. She can be contacted at sujinkim@uky.edu.
| Find other papers on this subject | ||
Information
You are invited to participate in a study about human biological repositories. Advances in genetic studies have led to significant uses of human biological samples. In this study about 100 individuals who are 18 and older will be asked to complete an online survey. The purpose of this study is to assess information requirements of human biological samples.
Benefits
This study will help tissue-centric users and information professionals including science, technology, and medical librarians as well as system developer alike understand specific requirements of tissue-centric users.
Potential risks
There are no foreseeable risks associated with this study. However, if you feel uncomfortable answering the survey questions, you may choose to skip a question or withdraw from the study at any time.
Confidentiality
The information collected form this survey will be accessible only to the researcher (PI: Sujin Kim). No personal identifiers will be used, and information will be presented in aggregate form, text delimited file only accessible to the researcher. No individual machine IP will be obtained. Random numbers will be assigned to obtain the aggregated data once the survey is submitted to the secure server. The survey will be collected on a server with SSL (Secure Sockets Layer) capabilities, which is one of the best providers of Internet security available, but there is always a risk that a third party may intercept the survey answers.
Contact
If you have questions, you may contact the principal investigator, Sujin Kim, at the University of Kentucky, School of Library and Information Science, 518 King Library, Lexington, KY, 40506, or sujinkim@uky.edu. If you have any questions about your rights as a volunteer in this research, contact XXXXX, Research Compliance Officer (XXXX@uky.edu) in the Office of Research Integrity at the University of Kentucky at XXX-XXX-XXXX or toll free at 1-800-XXX-XXXX.
Participation
Your participation in this study is voluntary; you may refuse to participate without penalty. If you withdraw from the study before data collection is completed, your data will be destroyed.
Consent
I have read this form, am 18 years of age or older, and agree to take part in this study.____ Yes
____ No
1. Year of birth (e.g., 1950): ____________
2. Gender: a. Male ____ b. Female ____
3. Highest degree earned: a. College student b. BS c. MS d. Ph.D. e. MD. F. Other _______
4. Your current institution:
a. Government
b. Non-profit research center
c. University hospital
d. Industry-sponsored research institution
e. University-based research center
f. Other: ____________________
5. Years of work experience: __________years
6. Specialty:
a. Molecular biology
b. Genetics
c. Pathology
d. Biochemistry
e. Public health
f. Biostatistics
g. Administration
h. Other________________________
7. Do you require human biological samples? Yes ______ No _______
8. Do you require animal and plant samples? Yes ______ No _______
9. Have you ever used human bio-repositories? Yes ______ No _______
10. Have you ever worked in human bio-repositories? Yes ______ No _______
11. Types of bio-repository work (please answer if you say 'Yes' in question 10.)
a. Sample collection
b. Sample processing
c. Sample storage
d. Sample distribution
e. Data management
f. Ethnical, legal, and procedural issue management
g. Other _________________________
12. What type(s) of samples do you require for your work?
1) Molecular
| a. cDNA/DNA | e. RNA, cytoplasmic | |
| b. Not specified | f. RNA, nuclear | |
| c. Protein | g. RNA, poly-A enriched | |
| d. RNA |
2) Cell
| a. Cryopreserved cells | c. Frozen cell block | |
| b. Fixed cell block | d. Frozen cell pellet |
3) Fluid
| a. Feces | k. Body cavity fluid | |
| b. Sweat | l. Milk | |
| c. Synovial fluid | m. Pericardial fluid | |
| d. Bile | n. Lavage | |
| e. Cerebrospinal fluid | o. Whole blood | |
| f. Amniotic fluid | p. Vitreous fluid | |
| g. Serum | q. Gastric fluid | |
| h. Whole bone marrow | r. Bone marrow plasma | |
| i. Saliva | s. Urine | |
| j. Plasma |
4) Tissue
| a. Frozen tissue | e. Fixed tissue slide | |
| b. Microdissected | f. Frozen tissue block | |
| c. Fresh tissue | g. Fixed tissue block | |
| d. Frozen tissue slide | h. Fixed tissue |
5) Other: ______________________________
13. What processing techniques do you require for your samples?
| a. PCR | j. IHC | |
| b. FISH | k. Mass spec. | |
| c. CGH | l. Westerns | |
| d. Sequencing | m. 1D/2D Gels | |
| e. ENPs | n. Ultra structure | |
| f. Microarrays | o. Microscopy (Light and EM) | |
| g. RT-PCR | p. Subcellular localization | |
| h. Northern | q. Other _________________ | |
| i. In situ hybridization |
14. What are the storage format(s) in which you would like to keep your samples?
| a. Formalin | e. Paraffin | |
| b. Fresh-frozen | f. Vials | |
| c. Frozen | g. Other _________________ | |
| d. OCT |
Amount: ________________ Unit of measurement: _____________
16. How often do you require samples? ______________times/year
17. Where do you normally seek bio-samples?
| a. Pathology department in university hospitals | e. Academic or non-profit research laboratories | |
| b. Sample donors directly through study protocol | f. Industrial/profit research laboratories | |
| c. Korean bio-repositories | g. Colleagues | |
| d. Oversee bio-repositories | h. Other ____________________. |
18. How much are you willing to spend to acquire samples for your work? __________ KRW
19. Where is your primary funding source?| a. Korean Science and Technology Research Foundation | e. Industry | |
| b. Ministry of Science and Technology | f. Non-profit research foundation | |
| c. Ministry of Health and Welfare | g. Private resource | |
| d. University hospital | h. Other ______________________ |
20. What anatomic region is your primary interest for which you require samples?
| a. Cardiovascular and circulatory system | h. Nervous system | |
| b. Digestive system | i. Respiratory system | |
| c. Embryonic structures | j. Sense organs | |
| d. Endocrine system | k. Stomatognathic system | |
| e. Hemic and immune systems | l. Urogenital system | |
| f. Integumentary system | m. Other _______________ | |
| g. Musculoskeletal system |
21. What are your searching criteria if you specify your sample requirement?
| a. By anatomic sites | g. By tissue sources (e.g., surgical or autopsy etc.) | |
| b. Either by normal or diseased | h. By demographic condition, (e.g., age, race, gender or other limiting characteristics) | |
| c. By disease | i. By amount of tissue(s) and minimum to maximum size or dimension | |
| d. By primary or metastatic | j. By total number of specimens needed | |
| e. By matched normal tissue from the same patient | k. By specimen preparation and preservation methods (e.g., fresh or frozen or fixed etc.) | |
| f. By specific treatment(s) performed on patient (e.g., radiation or chemo or hormone etc.) | l. Other ________________________ |
22. What diseases are of primary interest to you?
| a. Digestive system diseases | ,td>h. Endocrine system diseases | |
| b. Musculoskeletal diseases | i. Urogenital diseases | |
| c. Congenital, hereditary, and neonatal diseases and abnormalities | j. Respiratory tract diseases | |
| d. Cardiovascular diseases | k. Skin and connective tissue diseases | |
| e. Immune system diseases | l. Nervous system diseases | |
| f. Neoplasms | m. Other ______________________ | |
| g. Hemic and lymphatic diseases |
23. What are your publication outlets to report your work using bio-samples?
______________________________________________
24. What professional conferences do you regularly attend?
_____________________________________________
How much do you agree or disagree that the following medical and personal information should accompany human biological samples with 1 representing rarely useful and 5 representing extremely useful?
| 25. Names | 58. Operation history | |
| 26. Address | 59. Preoperative history | |
| 27. Dates | 60. Family health history | |
| 28. Telephone | 61. Clinical blood testing result | |
| 29. Fax | 62. Blood chemistry | |
| 30. E-mail | 63. Pulmonary function test | |
| 31. Social security number | 64. Electrolyte | |
| 32. Medical record number | 65. Virus | |
| 33. Health plan beneficiary numbers | 66. Gene expression | |
| 34. Insurance account numbers | 67. Biomarker | |
| 35. Certificate or license numbers | 68. Organ site | |
| 36. Vehicle identifiers and serial numbers | 69. How biological samples are processed | |
| 37. Device ID and serial numbers | 70. Laterality (which part of organ was taken, left, right, upper, lower, etc.) | |
| 38. Web address | 71. Pathologic diagnosis | |
| 39. Internet protocol address | 72. Radiological findings | |
| 40. Biometric identifier | 73. Treatment history | |
| 41. Photo | 74. World Health Organization (WHO) histologic grading | |
| 42. Unique id (e.g., university number) | 75. Progression or recurrence | |
| 43. Patient age | 76. Pathologic stage | |
| 44. Education | 77. Pathologic grade | |
| 45. Gender | 78. Institution where the sample was collected | |
| 46. Marital status | 79. Date and time collected | |
| 47. Occupation | 80. Date and time stored | |
| 48. Occupational-environmental | 81. Sample processed staff | |
| 49. Income | 82. Tumor or non-tumor | |
| 50. Donorís residence | 83. Sample type | |
| 51. Smoking | 84. Sample processing method | |
| 52. Nutrition | 85. Sample amount available | |
| 53. Alcohol | 86. How much dispensed | |
| 54. OBGYN history | 87. Where dispensed | |
| 55. Drug history | 88. Study results for dispensed samples | |
| 56. Physical examination (weight/height) | 89. Publication information for the sample that are dispensed for other users | |
| 57. Clinical diagnosis | 90. Other _______________________ |
© the author, 2010. Last updated: 12 March, 2010 |